Figure 1
UMAP on 7 MOFA factors
## UMAP
plot.umap.data <- plot_dimred(mofa, method="UMAP", color_by = "condition",stroke = 0.001, dot_size =1, alpha = 0.2, return_data = T)
plot.umap.all <- ggplot(plot.umap.data, aes(x=x, y = y, fill = color_by))+
geom_point(size = 0.8, alpha = 0.6, shape = 21, stroke = 0) +
theme_half_open() +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg",)+
theme(legend.position="none")+
scale_x_reverse()+
scale_y_reverse()+
add.textsize +
labs(title = "UMAP on MOFA+ factors shows minute and \nhour timescale signal transductions", x = "UMAP 1", y = "UMAP 2")
## UMAP legend
legend.umap <- get_legend( ggplot(plot.umap.data, aes(x=x, y = y, fill = color_by))+
geom_point(size = 2, alpha = 1, shape = 21, stroke = 0) +
theme_half_open() +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)",)+
add.textsize +
labs(title = "UMAP on MOFA+ factors shows minute and \nhour timescale signal transductions", x = "UMAP 1", "y = UMAP 2"))
legend.umap <- as_ggplot(legend.umap)plot_fig1_umap <- plot_grid(plot.umap.all,legend.umap, labels = c(""), label_size = 10, ncol = 2, rel_widths = c(1, 0.2))
ggsave(plot_fig1_umap, filename = "output/paper_figures/Fig1_UMAP.pdf", width = 68, height = 62, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot_fig1_umap, filename = "output/paper_figures/Fig1_UMAP.png", width = 68, height = 62, units = "mm", dpi = 300)
plot_fig1_umapFigure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements
Suppl PCA & WNN
seu_combined_selectsamples <- RunPCA(seu_combined_selectsamples,assay = "SCT.RNA", features = genes.variable, verbose = FALSE, ndims.print = 0, reduction.name = "pca.RNA")
seu_combined_selectsamples <- RunPCA(seu_combined_selectsamples, assay = "PROT", features = proteins.all, verbose = FALSE, ndims.print = 0,reduction.name = "pca.PROT")## PCA analysis
plot.PCA.RNA <- DimPlot(seu_combined_selectsamples, reduction = "pca.RNA", group.by = "condition", pt.size =0.2) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
labs(title = "RNA PCA separates cells \nat hour timescale", x= "RNA PC 1", y = " RNA PC 2") +
add.textsize +
theme(legend.position = "none")
plot.PCA.PROT <- DimPlot(seu_combined_selectsamples, reduction = "pca.PROT", group.by = "condition", pt.size = 0.2) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
labs(title = "(Phospho)protein PCA separates cells\nat minutes timescale", x= "Protein PC 1", y = "Protein PC 2") +
add.textsize +
# scale_x_reverse()+
theme(legend.position = "none")
legend <- get_legend(DimPlot(seu_combined_selectsamples, reduction = "pca.RNA", group.by = "condition", pt.size =0.1) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
add.textsize)
legend <- as_ggplot(legend)
PCA.PROTPC1.data <- data.frame(rank = 1:80,
protein = names(sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1])),
weight.PC1 = sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1]),
highlights = c(names(sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1]))[1:4],rep("",76))
)
PCA.PROTPC1.data <- PCA.PROTPC1.data %>%
mutate(highlights = as.character(highlights)) %>%
mutate(highlights = case_when(as.character(highlights) == "p-Syk" ~ "p-SYK",
as.character(highlights) == "p-Src" ~ "p-SRC",
TRUE ~ highlights))
plot.PCA.PROTweightsPC1 <- ggplot(PCA.PROTPC1.data, aes(x=weight.PC1, y = rank, label = highlights)) +
geom_point(size=0.1) +
labs(title = "Top 4 protein loadings \n", x= "Weight Protein PC1") +
geom_point(color = ifelse(PCA.PROTPC1.data$highlights == "", "grey50", "red")) +
geom_text_repel(size = 1.5, segment.size = 0.25)+
theme_half_open()+
add.textsize +
theme(axis.title.y=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
) +
scale_x_reverse()+
scale_y_reverse()
PCA.RNAPC1.data <- data.frame(rank = 1:length(genes.variable),
RNA = names(sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1])),
weight.PC1 = sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1]),
highlights = c(rep("",(length(genes.variable)-4)),names(sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1]))[(length(genes.variable)-3):length(genes.variable)])
)
## Loadings RNA
plot.PCA.RNAweightsPC1 <- ggplot(PCA.RNAPC1.data, aes(x=weight.PC1, y = rank, label = highlights)) +
geom_point(size=0.1) +
labs(title = "Top 4 RNA loadings \n", x= "Weight RNA PC1") +
geom_point(color = ifelse(PCA.RNAPC1.data$highlights == "", "grey50", "red")) +
geom_text_repel(size = 1.5, segment.size = 0.25)+
theme_half_open()+
add.textsize +
theme(axis.title.y=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
## ridgeplots
PC1.data <- data.frame(sample = rownames(seu_combined_selectsamples@reductions$pca.PROT@cell.embeddings),
PC1_PROT = seu_combined_selectsamples@reductions$pca.PROT@cell.embeddings[,1],
PC1_RNA = seu_combined_selectsamples@reductions$pca.RNA@cell.embeddings[,1]) %>%
left_join(meta.allcells)
plot_ridge_PC1Prot <- ggplot(PC1.data, aes(x = PC1_PROT, y = condition, fill = condition)) +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)")+
geom_density_ridges2() +
scale_y_discrete(labels = labels, name = "Time aIg (minutes)")+
scale_x_continuous(name = "PC1 Proteins") +
theme_half_open() +
add.textsize+
theme(legend.position = "none")
plot_ridge_PC1RNA <- ggplot(PC1.data, aes(x = PC1_RNA, y = condition, fill = condition)) +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)")+
geom_density_ridges2() +
scale_y_discrete(labels = labels, name = "Time aIg (minutes)")+
scale_x_continuous(name = "PC1 RNA") +
theme_half_open() +
add.textsize+
theme(legend.position = "none")seu_combined_selectsamples <- FindMultiModalNeighbors(
seu_combined_selectsamples, reduction.list = list("pca.RNA", "pca.PROT"),
dims.list = list(c(1:7), c(1:7)), modality.weight.name = "RNA.weight"
)
seu_combined_selectsamples <- RunUMAP(seu_combined_selectsamples, nn.name = "weighted.nn", reduction.name = "wnn.umap", reduction.key = "wnnUMAP_")
WNN.Dimplot <- DimPlot(seu_combined_selectsamples, reduction = 'wnn.umap', label = FALSE, repel = TRUE, label.size = 2.5) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
labs(title = "WNN UMAP similar to \n MOFA+ based UMAP", x= "wnnUMAP 1", y = "wnnUMAP 2") +
add.textsize +
# scale_x_reverse()+
theme(legend.position = "right")Factorvalues <- data.frame(get_factors(mofa)[1])[,1:4]
colnames(Factorvalues) <- c(paste0("Factor", 1:4))
seu_combined_selectsamples <- AddMetaData(seu_combined_selectsamples, Factorvalues)
plot.wnn.Factor1 <- FeaturePlot(seu_combined_selectsamples, reduction = "wnn.umap", features = "Factor1") +
scale_color_gradient2(name = "Factor 1 \nvalue") +
labs(title = "MOFA+ Factor 1 & 3 overlap \nwith WNN UMAP", x= "wnnUMAP 1", y = "wnnUMAP 2") +
add.textsize +
theme(legend.position = "right")
plot.wnn.Factor3 <- FeaturePlot(seu_combined_selectsamples, reduction = "wnn.umap", features = "Factor3") +
scale_color_gradient2(name = "Factor 3 \nvalue") +
labs(title = "", x= "wnnUMAP 1", y = "wnnUMAP 2") +
add.textsize +
# scale_x_reverse()+
theme(legend.position = "right")
plot.wnn.Factor1 <- FeaturePlot(seu_combined_selectsamples, reduction = "wnn.umap", features = "Factor1") +
scale_color_gradient2(name = "Factor 1 \nvalue") +
labs(title = "MOFA+ Factor 1 & 3 overlap \nwith WNN UMAP", x= "wnnUMAP 1", y = "wnnUMAP 2") +
add.textsize +
theme(legend.position = "right")
plot.wnn.pSYK <- FeaturePlot(seu_combined_selectsamples, reduction = "wnn.umap", features = "p-Syk", slot = "scale.data") +
scale_color_gradient2(name = "p-SYK \nscaled counts") +
labs(title = "\n", x= "wnnUMAP 1", y = "wnnUMAP 2") +
add.textsize +
# scale_x_reverse()+
theme(legend.position = "right")Fig1.pca <- plot_grid(plot.PCA.PROT,plot.PCA.RNA,legend, plot_ridge_PC1Prot, plot_ridge_PC1RNA,NULL, plot.PCA.PROTweightsPC1, plot.PCA.RNAweightsPC1,NULL,WNN.Dimplot,plot.wnn.pSYK,NULL,plot.wnn.Factor1,plot.wnn.Factor3, labels = c(panellabels[1:2], "",panellabels[3:4],"",panellabels[5:6],"",panellabels[7:8],"",panellabels[9:10]), label_size = 10, ncol = 3, rel_widths = c(0.9,0.9,0.2,0.9,0.9), rel_heights = c(1,0.8,1.3,1))
ggsave(filename = "output/paper_figures/Suppl_PCA_aIg_wnn.pdf", width = 143, height = 226, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(filename = "output/paper_figures/Suppl_PCA_aIg_wnn.png", width = 143, height = 226, units = "mm", dpi = 300)
Fig1.pcaSupplementary Figure. PCA analysis on RNA and Protein datasets separately.
Suppl MOFA model properties
## Variance per factor
plot.variance.perfactor.all <- plot_variance_explained(mofa, x="factor", y="view") +
add.textsize +
labs(title = "Variance explained by each factor per modality")
## variance total
plot.variance.total <- plot_variance_explained(mofa, x="view", y="factor", plot_total = T)
plot.variance.total <- plot.variance.total[[2]] +
add.textsize +
labs(title = "Total \nvariance") +
geom_text(aes(label=round(R2,1)), vjust=1.6, color="white", size=2.5)
## Significance correlation covariates
plot.heatmap.pval.covariates <- as.ggplot(correlate_factors_with_covariates(mofa,
covariates = c("time"),
factors = 1:mofa@dimensions$K,
fontsize = 7,
cluster_row = F,
cluster_col = F
))+
add.textsize +
theme(axis.text.y=element_blank(),
axis.text.x=element_blank(),
plot.title = element_text(size=7, face = "plain"),
)## Factor values over time
plot.violin.factorall <- plot_factor(mofa,
factor = c(2,4:mofa@dimensions$K),
color_by = "condition",
dot_size = 0.2, # change dot size
dodge = T, # dodge points with different colors
legend = F, # remove legend
add_violin = T, # add violin plots,
violin_alpha = 0.9 # transparency of violin plots
) +
add.textsize +
scale_color_manual(values=c(colorgradient6_manual2, labels = c(labels), name = "Time aIg")) +
scale_fill_manual(values=c(colorgradient6_manual2, labels = c(labels), name = "Time aIg"))+
labs(title = "Factor values per time-point of additional factors not correlating with time." ) +
theme(axis.text.x=element_blank())
## Loadings factors stable protein
plot.rank.PROT.2.4to7 <- plot_weights(mofa,
view = "PROT",
factors = c(1:mofa@dimensions$K),
nfeatures = 3,
text_size = 1.5
) +
add.textsize +
labs(title = "Top 3 Protein loadings per factor") +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
## Loadings factors stable protein
plot.rank.RNA.2.4to7 <- plot_weights(mofa,
view = "RNA",
factors = c(1:mofa@dimensions$K),
nfeatures = 3,
text_size = 1.5
) +
add.textsize +
labs(title = "Top 3 RNA loadings per factor") +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
## correlation time and factors
plot.correlation.covariates <- correlate_factors_with_covariates(mofa,
covariates = c("time"),
factors = mofa@dimensions$K:1,
plot = "r"
)plot.correlation.covariates <- ggcorrplot(plot.correlation.covariates, tl.col = "black", method = "square", lab = TRUE, ggtheme = theme_void, colors = c("#11304C", "white", "#11304C"), lab_size = 2.5) +
add.textsize +
labs(title = "Correlation of \nfactors with \ntime of treatment", y = "") +
scale_y_discrete(labels = "") +
coord_flip() +
theme(axis.text.x=element_text(angle =0,hjust = 0.5),
axis.text.y=element_text(size = 5),
legend.position="none",
plot.title = element_text(hjust = 0.5))Fig.1.suppl.mofa.row1 <- plot_grid(plot.variance.perfactor.all, plot.variance.total,NULL, plot.heatmap.pval.covariates, labels = c(panellabels[1:3]), label_size = 10, ncol = 4, rel_widths = c(1.35, 0.3,0.25,0.38))
Fig.1.suppl.mofa.row2 <- plot_grid(plot.violin.factorall,legend.umap, labels = panellabels[4], label_size = 10, ncol = 2, rel_widths = c(1,0.1))
Suppl_mofa <- plot_grid(Fig.1.suppl.mofa.row1, Fig.1.suppl.mofa.row2, plot.rank.PROT.2.4to7, plot.rank.RNA.2.4to7, labels = c("","", panellabels[5:6]),label_size = 10, ncol = 1, rel_heights = c(1.45,1,1.1,1.1))
ggsave(Suppl_mofa, filename = "output/paper_figures/Fig2.Suppl_MOFAaIg.pdf", width = 183, height = 220, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(Suppl_mofa, filename = "output/paper_figures/Fig2.Suppl_MOFAaIg.png", width = 183, height = 220, units = "mm", dpi = 300)
Suppl_mofaSupplementary Figure. MOFA model additional information
Figure 2
Prepare main panels
meta.allcells <- seu_combined_selectsamples@meta.data %>%
mutate(sample = rownames(seu_combined_selectsamples@meta.data))
proteindata <- as.data.frame(t(seu_combined_selectsamples@assays$PROT@scale.data)) %>%
mutate(sample = rownames(t(seu_combined_selectsamples@assays$PROT@scale.data))) %>%
left_join(meta.allcells)
MOFAfactors<- as.data.frame(mofa@expectations$Z) %>%
mutate(sample = rownames(as.data.frame(mofa@expectations$Z)[,1:mofa@dimensions$K]))
MOFAfactors <- left_join(as.data.frame(MOFAfactors), meta.allcells)
weights.prot <- get_weights(mofa, views = "PROT",as.data.frame = TRUE)
topnegprots.factor1 <- weights.prot %>%
filter(factor == "Factor1" & value <= 0) %>%
arrange(value)
topposprots.factor3 <- weights.prot %>%
filter(factor == "Factor3" & value >= 0) %>%
arrange(-value)
weights.RNA <- get_weights(mofa, views = "RNA",as.data.frame = TRUE)## violin plots phospho-proteins highlighted in main
f.violin.fact <- function(data , protein){
ggplot(subset(data) , aes(x = as.factor(condition), y =get(noquote(protein)))) +
annotate("rect",
xmin = 5 - 0.45,
xmax = 6 + 0.6,
ymin = -4.5, ymax = 4.5, fill = "lightblue",
alpha = .4
)+
geom_hline(yintercept=0, linetype='dotted', col = 'black')+
geom_violin(alpha = 0.9,aes(fill = condition))+
geom_jitter(width = 0.05,size = 0.1, fill = "black", shape = 21)+
stat_summary(fun=median, geom="point", shape=95, size=2, inherit.aes = T, position = position_dodge(width = 0.9), color = "red")+
theme_few()+
ylab(paste0(protein)) +
scale_x_discrete(labels = labels, expand = c(0.1,0.1), name = "Time aIg (minutes)") +
scale_y_continuous(expand = c(0,0), name = strsplit(protein, split = "\\.")[[1]][2]) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",)+
scale_fill_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",) +
add.textsize +
theme(#axis.title.x=element_blank(),
#axis.text.x=element_blank(),
#axis.ticks.x=element_blank(),
legend.position="none")
}
factors_toplot <- c(colnames(MOFAfactors)[c(1,3)])
for(i in factors_toplot) {
assign(paste0("plot.factor.", i), f.violin.fact(data = MOFAfactors,protein = i))
}
legend.violinfactor <- as_ggplot( get_legend( ggplot(subset(MOFAfactors) , aes(x = as.factor(condition), y =get(noquote(factors_toplot[1])))) +
annotate("rect",
xmin = 5 - 0.45,
xmax = 6 + 0.6,
ymin = -4.5, ymax = 4.5, fill = "lightblue",
alpha = .4
)+
geom_hline(yintercept=0, linetype='dotted', col = 'black')+
geom_violin(alpha = 0.9,aes(fill = condition))+
geom_jitter(width = 0.05,size = 0.1, aes(col = condition), fill = "black", shape = 21)+
stat_summary(fun=median, geom="point", shape=23, size=2, inherit.aes = T, position = position_dodge(width = 0.9), color = "black")+
theme_few()+
ylab(paste0(factors_toplot[1])) +
scale_x_discrete(labels = labels, expand = c(0.1,0.1), name = "Time aIg (minutes)") +
scale_y_continuous(expand = c(0,0), name = strsplit(factors_toplot[1], split = "\\.")[[1]][2]) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg \n(minutes)",)+
scale_fill_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg \n(minutes)",) +
add.textsize ) )
plot.violin.factor1 <- plot.factor.group1.Factor1 +
scale_y_reverse(expand = c(0,0), name = "Factor 1 value")+
annotate(geom="text", x=1.5, y=-4.3, label="minutes", size = 2,
color="grey3") +
annotate(geom="text", x=5.5, y=-4.3, label="hours", size = 2,
color="grey3") +
labs(title = "Factor 1 captures \nminute timescale signal transduction")
plot.violin.factor3 <- plot.factor.group1.Factor3 +
annotate(geom="text", x=5.5, y=4.3, label="hours", size = 2,
color="grey3")+
annotate(geom="text", x=1.5, y=4.3, label="minutes", size = 2,
color="grey3") +
scale_y_continuous(expand = c(0,0), name = "Factor 3 value")+
labs(title = "Factor 3 captures \nhour timescale signal transduction")
#### protein Loadings factor 1
list.bcellpathway.protein <- c("p-CD79a", "p-SYK", "p-SRC", "p-ERK1/2", "p-PLC-y2", "p-BLNK","p-PLC-y2Y759","p-PKC-b1", "p-p38", "p-AKT", "p-S6", "p-TOR", "CD79a") # , "p-PLC-y2", "p-PKC-b1", "p-IKKa/b","p-JNK", "p-p38", "p-p65","p-Akt", "p-S6", "p-TOR"
list.top20 <- topnegprots.factor1$feature[1:20]
## protein factor 1
plotdata.rank.PROT.1 <-plot_weights(mofa,
view = "PROT",
factors = c(1),
nfeatures = 15,
text_size = 1,
manual = list(list.top20, list.bcellpathway.protein, "p-JAK1" ),
color_manual = list("grey","blue4","red"),
return_data = TRUE
)
plotdata.rank.PROT.1 <- plotdata.rank.PROT.1 %>%
mutate(Rank = 1:80,
Weight = value,
colorvalue = ifelse(labelling_group == 2 & value <= -0.2,"grey", ifelse(labelling_group == 3 & value <= -0.2, "blue4", ifelse(labelling_group == 4 & value <= -0.2, "red", "grey"))),
highlights = ifelse(labelling_group >= 1 & value <= -0.25, as.character(feature), "")
)
plot.rank.PROT.1 <- ggplot(plotdata.rank.PROT.1, aes(x=Weight, y = Rank, label = highlights)) +
labs(title = "(Phospho)proteins: <p><span style='color:blue4'>BCR </span>&<span style='color:red'> JAK1</span> signaling", #<span style='color:blue4'>BCR signaling</span> and <span style='color:red'>p-JAK1</span>
x= "Factor 1 loading value",
y= "Factor 1 loading rank") +
geom_point(size=0.1, color =plotdata.rank.PROT.1$colorvalue, size =1) +
geom_text_repel(size = 2,
segment.size = 0.2,
color =plotdata.rank.PROT.1$colorvalue,
nudge_x = 1 - plotdata.rank.PROT.1$Weight,
direction = "y",
hjust = 1,
segment.color = "grey50")+
theme_few()+
add.textsize +
scale_y_continuous(trans = "reverse") +
add.textsize +
theme(
plot.title = element_markdown()
) +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)+
xlim(c(-1,1))
## violin plots phospho-proteins highlighted in main
f.violin.prot <- function(data , protein){
ggplot(subset(data) , aes(x = as.factor(condition), y =get(noquote(protein)))) +
annotate("rect",
xmin = 5 - 0.45,
xmax = 6 + 0.6,
ymin = -5.5, ymax = 5.5, fill = "lightblue",
alpha = .4
)+
geom_violin(alpha = 0.9,aes(fill = condition))+
geom_jitter(width = 0.05,size = 0.1, color = "black")+
stat_summary(fun=median, geom="point", shape=95, size=2, inherit.aes = T, position = position_dodge(width = 0.9), color = "red")+
theme_few()+
ylab(paste0(protein)) +
scale_x_discrete(labels = labels, expand = c(0.1,0.1), name = "Time aIg (minutes)") +
scale_y_continuous(expand = c(0,0), name = protein) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",)+
scale_fill_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",) +
add.textsize +
theme(#axis.title.x=element_blank(),
#axis.text.x=element_blank(),
#axis.ticks.x=element_blank(),
legend.position="none")
}
proteins_toplot <- c("p-CD79a","p-Syk","p-JAK1", "IgM")
for(i in proteins_toplot) {
assign(paste0("plot.violin.", i), f.violin.prot(data = proteindata ,protein = i))
}
## Fig 2 bottom row panels
`plot.violin.p-CD79a` <- `plot.violin.p-CD79a` +
labs(title = "BCR signaling pathway and \nJAK1 activation")+
add.textsize+
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),)
`plot.violin.p-Syk` <- `plot.violin.p-Syk` +
add.textsize+
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),) +
scale_y_continuous(name = "p-SYK")
`plot.violin.p-JAK1` <-`plot.violin.p-JAK1` +
add.textsize +
labs(y = "<span style='color:red'>p-JAK1</span>") +
theme(axis.title.y = element_markdown()) Enrichment analysis of Factor 3 positive loadings
topposrna.factor3 <- weights.RNA %>%
filter(factor == "Factor3" & value >= 0) %>%
arrange(-value)
rownames(topposrna.factor3) <- topposrna.factor3$feature
### Convert Gene-names to gene IDs (using 'org.Hs.eg.db' library)
topposrna.factor3 <- topposrna.factor3 %>%
mutate(ENTREZID = mapIds(org.Hs.eg.db, as.character(topposrna.factor3$feature), 'ENTREZID', 'SYMBOL'))
listgenes.factor3 <- topposrna.factor3$value
names(listgenes.factor3) <- topposrna.factor3$ENTREZID
go.pb.fct3.pos <- enrichGO(gene = topposrna.factor3$feature,
OrgDb = org.Hs.eg.db,
keyType = 'SYMBOL',
ont = "BP",
pAdjustMethod = "BH")
go.pb.fct3.pos <- simplify(go.pb.fct3.pos, cutoff=0.6, by="p.adjust", select_fun=min)
go.pb.fct3.pos.dplyr <- mutate(go.pb.fct3.pos, richFactor = Count / as.numeric(sub("/\\d+", "", BgRatio)))
## plot main panel
plot.enriched.go.bp.factor3 <- ggplot(go.pb.fct3.pos.dplyr, showCategory = 10,
aes(-log10(p.adjust), fct_reorder(Description, -log10(p.adjust)))) + geom_segment(aes(xend=0, yend = Description)) +
geom_point(aes(size = Count)) +
scale_color_viridis_c(guide=guide_colorbar(reverse=TRUE)) +
scale_size_continuous(range=c(0.5, 3)) +
theme_minimal() +
add.textsize +
xlab("-log adj pval") +
ylab(NULL) +
ggtitle("Enriched Biological Processes") +
theme(legend.position="none")
legend.plot.enriched.go.bp.factor3 <- as.ggplot(get_legend(ggplot(go.pb.fct3.pos.dplyr, showCategory = 10,
aes(-log10(p.adjust), fct_reorder(Description, -log10(p.adjust)))) + geom_segment(aes(xend=0, yend = Description)) +
geom_point(aes(size = Count)) +
scale_color_viridis_c(guide=guide_colorbar(reverse=TRUE)) +
scale_size_continuous(range=c(0.5, 3)) +
theme_minimal() +
add.textsize +
xlab("-log adj pval") +
ylab(NULL) +
ggtitle("Enriched Biological Processes") +
theme(legend.position="right")))
## Plot supplementary
plot.enriched.go.bp.factor3_50 <- ggplot(go.pb.fct3.pos.dplyr, showCategory = 50,
aes(-log10(p.adjust), fct_reorder(Description, -log10(p.adjust)))) + geom_segment(aes(xend=0, yend = Description)) +
geom_point(aes(size = Count)) +
scale_color_viridis_c(guide=guide_colorbar(reverse=TRUE)) +
scale_size_continuous(range=c(0.5, 3)) +
theme_minimal() +
add.textsize +
xlab("-log adj pval") +
ylab(NULL) +
ggtitle("Enriched Biological Processes") message(c("Significance protein folding GO term: \n",go.pb.fct3.pos.dplyr@result[which(go.pb.fct3.pos.dplyr@result$Description == "protein folding"),"p.adjust"]))##For RNA loadings panels
topgeneset.fct3<- unlist(str_split(go.pb.fct3.pos.dplyr@result[1:10,"geneID"], pattern = "/"))
topgeneset.fct3 = bitr(topgeneset.fct3, fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
## RNA factor 1 loadings
plotdata.rank.RNA.3 <-plot_weights(mofa,
view = "RNA",
factors = c(3),
nfeatures = 5,
text_size = 1,
manual = list(topposrna.factor3$feature[c(1:5)] ,topgeneset.fct3$SYMBOL),
color_manual = list("grey","blue4"),
return_data = TRUE
)
plotdata.rank.RNA.3 <- plotdata.rank.RNA.3 %>%
mutate(Rank = 1:nrow(plotdata.rank.RNA.3),
Weight = value,
colorvalue = ifelse(labelling_group == 3 &value >= 0.25,"blue4", ifelse(labelling_group == 2&value >= 0.25, "grey2", "grey")),
highlights = ifelse(labelling_group >= 1&value >= 0.25, as.character(feature), "")
)
plot.rank.RNA.3 <- ggplot(plotdata.rank.RNA.3, aes(x=Weight, y = Rank, label = highlights)) +
labs(title = "Contributing genes <p><span style='color:blue4'><span style='color:blue4'>GO-term vesicle related </span> ", #<span style='color:blue4'>BCR signaling regulation (find GO term+list todo)</span>
x= "Factor 3 loading value",
y= "Factor 3 loading rank") +
geom_point(size=0.1, color =plotdata.rank.RNA.3$colorvalue) +
geom_text_repel(size = 2,
segment.size = 0.2,
color =plotdata.rank.RNA.3$colorvalue,
nudge_x = -1 - plotdata.rank.RNA.3$Weight,
direction = "y",
hjust = 0,
segment.color = "grey50")+
theme_few()+
add.textsize +
scale_y_continuous() +
add.textsize +
theme(
plot.title = element_markdown()
) +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank(),
axis.title.y = element_blank())+
xlim(c(-1,1))
#### Protein loadings factor 3
list.highlight.prot.fct3 <- c("p-p38", "p-AKT", "p-S6", "p-TOR", "XBP1_PROT", "p-STAT5")
list.top20 <- topnegprots.factor1$feature[1:20]
plotdata.rank.PROT.3 <-plot_weights(mofa,
view = "PROT",
factors = c(3),
nfeatures = 30,
text_size = 1,
manual = list(topposprots.factor3$feature[c(1:8)] ,list.highlight.prot.fct3),
color_manual = list("grey","blue4"),
return_data = TRUE
)
plotdata.rank.PROT.3 <- plotdata.rank.PROT.3 %>%
mutate(Rank = 1:80,
Weight = value,
colorvalue = ifelse(labelling_group == 3 &value >= 0.25,"blue4", ifelse(labelling_group == 2&value >= 0.4, "grey", "grey")),
highlights = ifelse(labelling_group >= 1&value >= 0.25, as.character(feature), "")
) %>%
mutate(highlights = case_when(as.character(highlights) == "XBP1_PROT" ~ "XBP1",
TRUE ~ highlights))
plot.rank.PROT.3 <- ggplot(plotdata.rank.PROT.3, aes(x=Weight, y = Rank, label = highlights)) +
labs(title = "Signaling implicated in <p><span style='color:blue4'>Unfolded protein response</span>",
x= "Factor 3 loading value",
y= "Factor 3 loading rank") +
geom_point(size=0.1, color =plotdata.rank.PROT.3$colorvalue, size =1) +
geom_text_repel(size = 2,
segment.size = 0.2,
color =plotdata.rank.PROT.3$colorvalue,
nudge_x = -1 - plotdata.rank.PROT.3$Weight,
direction = "y",
hjust = 0,
segment.color = "grey50")+
theme_few()+
add.textsize +
scale_y_continuous() +
add.textsize +
theme(
plot.title = element_markdown()
) +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)+
xlim(c(-1,1))
#### RNA loadings factor 1
plotdata.rank.RNA.1 <-plot_weights(mofa,
view = "RNA",
factors = c(1),
nfeatures = 2,
text_size = 1,
return_data = TRUE
)
plotdata.rank.RNA.1 <- plotdata.rank.RNA.1 %>%
mutate(Rank = 1:nrow(plotdata.rank.RNA.1),
Weight = value,
colorvalue = ifelse(labelling_group == 1,"blue4", ifelse(labelling_group == 1, "blue4", "grey")),
highlights = ifelse(labelling_group >= 1, as.character(feature), "")
)
plot.rank.RNA.1 <- ggplot(plotdata.rank.RNA.1, aes(x=Weight, y = Rank, label = highlights)) +
labs(title = " Contributing genes<p><span style='color:blue4'>BCR activation</span>",
x= "Factor 1 loading value",
y= "Factor 1 loading rank") +
geom_point(size=0.1, color =plotdata.rank.RNA.1$colorvalue) +
geom_text_repel(size = 2,
segment.size = 0.2,
color =plotdata.rank.RNA.1$colorvalue,
nudge_x = 1 - plotdata.rank.RNA.1$Weight,
direction = "y",
hjust = 1,
segment.color = "grey50") +
theme_few()+
add.textsize +
scale_y_continuous(trans = "reverse") +
add.textsize +
theme(
plot.title = element_markdown()
) +
theme(axis.text.y=element_blank(),
axis.ticks.y=element_blank(),
axis.title.y=element_blank()
)+
xlim(c(-1,1))
#### Violins genes
f.violin.rna <- function(data , protein){
ggplot(subset(data) , aes(x = as.factor(condition), y =get(noquote(protein)))) +
annotate("rect",
xmin = 5 - 0.45,
xmax = 6 + 0.6,
ymin = -3, ymax = 15, fill = "lightblue",
alpha = .4
)+
geom_violin(alpha = 0.9,aes(fill = condition))+
geom_jitter(width = 0.05,size = 0.1, color = "black")+
stat_summary(fun=median, geom="point", shape=95, size=2, inherit.aes = T, position = position_dodge(width = 0.9), color = "red")+
theme_few()+
ylab(paste0(protein)) +
scale_x_discrete(labels = labels, expand = c(0.1,0.1), name = "Time aIg (minutes)") +
scale_y_continuous(expand = c(0,0), name = protein) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",)+
scale_fill_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg ",) +
add.textsize +
theme(#axis.title.x=element_blank(),
#axis.text.x=element_blank(),
#axis.ticks.x=element_blank(),
legend.position="none")
}
rnadata <- as.data.frame(t(seu_combined_selectsamples@assays$SCT.RNA@scale.data)) %>%
mutate(sample = rownames(t(seu_combined_selectsamples@assays$SCT.RNA@scale.data))) %>%
left_join(meta.allcells)
enriched.geneset.posregBcell.fact1 <- c("NPM1", "NEAT1", "BTF3", "IGHM", "IGKC")
##Violin genes
for(i in enriched.geneset.posregBcell.fact1) {
assign(paste0("plot.violin.rna.", i), f.violin.rna(data = rnadata ,protein = i))
}
plot.violin.rna.NEAT1 <- plot.violin.rna.NEAT1 +
labs(title = "Expression of upregulated genes\n")+
add.textsize+
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),)
plot.violin.rna.NPM1 <- plot.violin.rna.NPM1 +
add.textsize+
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),)
Main figure 2
Fig2.row1.violinsprot <- plot_grid(`plot.violin.p-CD79a`, `plot.violin.p-Syk`, `plot.violin.p-JAK1`,labels = c(panellabels[4]), label_size = 10, ncol = 1, rel_heights = c(1.2,0.95,1.2))
Fig2.row1 <- plot_grid(plot.violin.factor1,plot.rank.PROT.1, NULL, Fig2.row1.violinsprot, labels = c(panellabels[1:3], ""), label_size = 10, ncol =4, rel_widths = c(0.8,0.55,0.5,0.8))
Fig2.row2.violinsrna <- plot_grid(plot.violin.rna.NEAT1,plot.violin.rna.NPM1, plot.violin.rna.BTF3,labels = "", label_size = 10, ncol = 1, rel_heights = c(1.2,0.95,1.2))
Fig2.row2 <- plot_grid(plot.violin.factor3,plot.rank.PROT.3,plot.rank.RNA.3,Fig2.row2.violinsrna, labels = c(panellabels[5:6], "", panellabels[8]), label_size = 10, ncol =4, rel_widths = c(0.8,0.55,0.5,0.8))
Fig2.row3 <- plot_grid(plot.enriched.go.bp.factor3,legend.plot.enriched.go.bp.factor3,NULL, labels = panellabels[7], label_size = 10, ncol =3, rel_widths = c(1.8,0.2,0.6))
plot_fig2 <- plot_grid(Fig2.row1, Fig2.row2,Fig2.row3, labels = c("", "", ""), label_size = 10, ncol = 1, rel_heights = c(1,1,0.55))
ggsave(plot_fig2, filename = "output/paper_figures/Fig2.pdf", width = 183, height = 183, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot_fig2, filename = "output/paper_figures/Fig2.png", width = 183, height = 183, units = "mm", dpi = 300)
plot_fig2Figure 2. Factor 1 and 3 exploration.
Suppl Enriched Fact 3
ggsave(plot.enriched.go.bp.factor3_50, filename = "output/paper_figures/Suppl_Enrichm_Fct3.pdf", width = 183, height = 183, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot.enriched.go.bp.factor3_50, filename = "output/paper_figures/Suppl_Enrichm_Fct3.png", width = 183, height = 183, units = "mm", dpi = 300)
plot.enriched.go.bp.factor3_50Supplementary Figure. Top 50 enriched gene-sets in postive loadings factor 3.
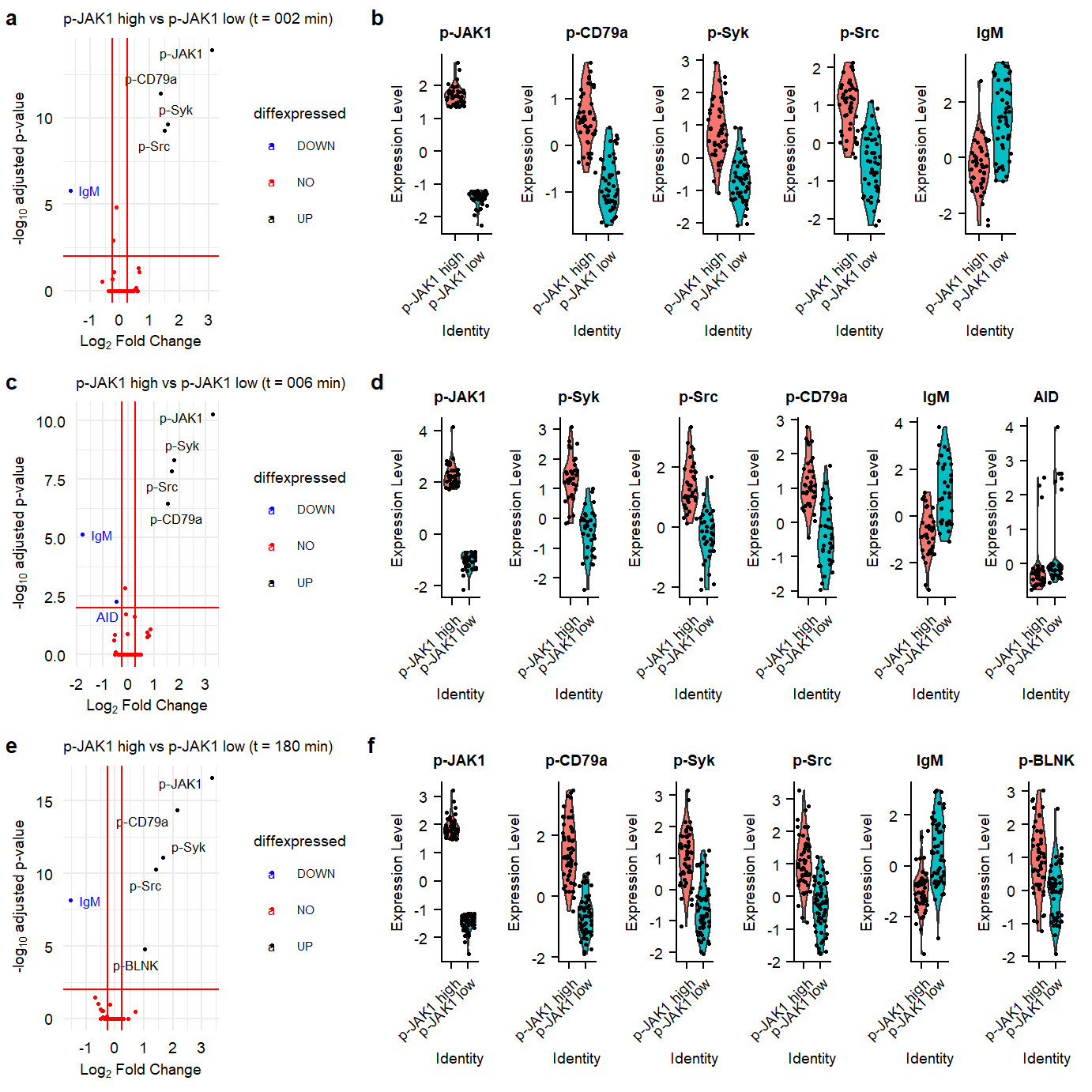
Suppl pJak1 High vs low
proteindata_counts <- as.data.frame(t(seu_combined_selectsamples@assays$PROT@scale.data)) %>%
mutate(sample = rownames(t(seu_combined_selectsamples@assays$PROT@scale.data)))
metadata.all <- as.data.frame(seu_combined_selectsamples@meta.data) %>%
mutate(sample = rownames((seu_combined_selectsamples@meta.data)))
proteindata_counts <- left_join(proteindata_counts, metadata.all)
toppJAK1 <- proteindata_counts %>%
group_by(time) %>%
top_frac(wt = `p-JAK1`, n = 0.05)
bottompJAK1 <- proteindata_counts %>%
group_by(time) %>%
top_frac(wt = `p-JAK1`, n = -0.05)
proteindata_counts <- proteindata_counts %>%
mutate(highlowpJAK1 = ifelse(sample %in% toppJAK1$sample, "p-JAK1 high", ifelse(sample %in% bottompJAK1$sample, "p-JAK1 low","middle")))
addmeta <- proteindata_counts[,c("highlowpJAK1", "sample")]
rownames(addmeta) <- proteindata_counts$sample
seu_combined_selectsamples <- AddMetaData(seu_combined_selectsamples, addmeta)
seu.JAK1.180 <- subset(seu_combined_selectsamples, condition == "180.aIg.contr" & highlowpJAK1 != "middle")
seu.JAK1.180 <- SetIdent(seu.JAK1.180, value = "highlowpJAK1")# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.180 <- FindMarkers(seu.JAK1.180,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "PROT", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.180 <- filter(markers.180, cluster == "p-JAK1 high")
#markers.180
# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.180.RNA <- FindAllMarkers(seu.JAK1.180,assay = "RNA", slot = "data", logfc.threshold = 0.3, return.thresh = 0.01, only.pos = T,min.pct = 0.1)
markers.180.RNA <- FindMarkers(seu.JAK1.180,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "SCT.RNA", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.180.RNA
markers.180$protein <-rownames(markers.180)
# add a column of NAs
markers.180$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.180$diffexpressed[markers.180$avg_diff > 0.25 & markers.180$p_val_adj < 0.01] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.180$diffexpressed[markers.180$avg_diff < -0.25 & markers.180$p_val_adj < 0.01] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.180$delabel <- NA
markers.180$delabel[markers.180$diffexpressed != "NO"] <- markers.180$protein[markers.180$diffexpressed != "NO"]
# Finally, we can organize the labels nicely using the "ggrepel" package and the geom_text_repel() function
# load library
# plot adding up all layers we have seen so far
plot.vulcano.180min <- ggplot(data=markers.180, aes(x=avg_diff , y=-log10(p_val_adj), col=diffexpressed, label=delabel)) +
geom_point(size=0.5) +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.01), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" adjusted p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 180 min)") &
add.textsize
sign.markers180 <- markers.180$protein[markers.180$avg_diff > 0.25 & markers.180$p_val_adj < 0.01 | markers.180$avg_diff < -0.25 & markers.180$p_val_adj < 0.01]
plot.vln.180min <- VlnPlot(seu.JAK1.180,assay = "PROT",slot = "scale.data", features = sign.markers180, group.by = "highlowpJAK1",ncol = 6, pt.size = 0.5) &
add.textsize
## RNA
markers.180.RNA$protein <-rownames(markers.180.RNA)
# add a column of NAs
markers.180.RNA$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.180.RNA$diffexpressed[markers.180.RNA$avg_diff > 0.25 & markers.180.RNA$p_val < 0.05] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.180.RNA$diffexpressed[markers.180.RNA$avg_diff < -0.25 & markers.180.RNA$p_val < 0.05] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.180.RNA$delabel <- NA
markers.180.RNA$delabel[markers.180.RNA$diffexpressed != "NO"] <- markers.180.RNA$protein[markers.180.RNA$diffexpressed != "NO"]
# plot adding up all layers we have seen so far
plot.vulcano.180min.RNA <- ggplot(data=markers.180.RNA, aes(x=avg_diff, y=-log10(p_val), col=diffexpressed, label=delabel)) +
geom_point() +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.05), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 180 min)") &
add.textsize
sign.markers180.RNA <- markers.180.RNA$protein[markers.180.RNA$avg_diff > 0.25 & markers.180.RNA$p_val < 0.05]
plot.vln.180min.RNA <- VlnPlot(seu.JAK1.180,assay = "RNA", features = sign.markers180.RNA[1:20], group.by = "highlowpJAK1",ncol = 10) &
add.textsize
plot_180min <- plot_grid(plot.vulcano.180min, plot.vln.180min, labels = panellabels[c(5,6)], label_size = 10, ncol = 2, rel_widths = c(1,2))
## 6 minutes
seu.JAK1.006 <- subset(seu_combined_selectsamples, condition == "006.aIg.contr" & highlowpJAK1 != "middle")
seu.JAK1.006 <- SetIdent(seu.JAK1.006, value = "highlowpJAK1")
# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.006 <- FindMarkers(seu.JAK1.006,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "PROT", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.006 <- filter(markers.006, cluster == "p-JAK1 high")
#markers.006
# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.006.RNA <- FindAllMarkers(seu.JAK1.006,assay = "RNA", slot = "data", logfc.threshold = 0.3, return.thresh = 0.01, only.pos = T,min.pct = 0.1)
markers.006.RNA <- FindMarkers(seu.JAK1.006,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "SCT.RNA", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.006.RNA
markers.006$protein <-rownames(markers.006)
# add a column of NAs
markers.006$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.006$diffexpressed[markers.006$avg_diff > 0.25 & markers.006$p_val_adj < 0.01] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.006$diffexpressed[markers.006$avg_diff < -0.25 & markers.006$p_val_adj < 0.01] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.006$delabel <- NA
markers.006$delabel[markers.006$diffexpressed != "NO"] <- markers.006$protein[markers.006$diffexpressed != "NO"]
# Finally, we can organize the labels nicely using the "ggrepel" package and the geom_text_repel() function
# load library
# plot adding up all layers we have seen so far
plot.vulcano.006min <- ggplot(data=markers.006, aes(x=avg_diff , y=-log10(p_val_adj), col=diffexpressed, label=delabel)) +
geom_point(size=0.5) +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.01), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" adjusted p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 006 min)") &
add.textsize
sign.markers006 <- markers.006$protein[markers.006$avg_diff > 0.25 & markers.006$p_val_adj < 0.01 | markers.006$avg_diff < -0.25 & markers.006$p_val_adj < 0.01]
plot.vln.006min <- VlnPlot(seu.JAK1.006,assay = "PROT",slot = "scale.data", features = sign.markers006, group.by = "highlowpJAK1",ncol = 6, pt.size = 0.5) &
add.textsize
markers.006.RNA$protein <-rownames(markers.006.RNA)
# add a column of NAs
markers.006.RNA$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.006.RNA$diffexpressed[markers.006.RNA$avg_diff > 0.25 & markers.006.RNA$p_val < 0.05] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.006.RNA$diffexpressed[markers.006.RNA$avg_diff < -0.25 & markers.006.RNA$p_val < 0.05] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.006.RNA$delabel <- NA
markers.006.RNA$delabel[markers.006.RNA$diffexpressed != "NO"] <- markers.006.RNA$protein[markers.006.RNA$diffexpressed != "NO"]
# Finally, we can organize the labels nicely using the "ggrepel" package and the geom_text_repel() function
# load library
# plot adding up all layers we have seen so far
plot.vulcano.006min.RNA <- ggplot(data=markers.006.RNA, aes(x=avg_diff, y=-log10(p_val), col=diffexpressed, label=delabel)) +
geom_point() +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.05), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 006 min)") &
add.textsize
sign.markers006.RNA <- markers.006.RNA$protein[markers.006.RNA$avg_diff > 0.25 & markers.006.RNA$p_val < 0.05]
plot.vln.006min.RNA <- VlnPlot(seu.JAK1.006,assay = "RNA", features = sign.markers006.RNA[1:20], group.by = "highlowpJAK1",ncol = 10) &
add.textsize
plot_006min <- plot_grid(plot.vulcano.006min, plot.vln.006min, labels = panellabels[c(3,4)], label_size = 10, ncol = 2, rel_widths = c(1,2))
## 2 minutes
seu.JAK1.002 <- subset(seu_combined_selectsamples, condition == "002.aIg.contr" & highlowpJAK1 != "middle")
seu.JAK1.002 <- SetIdent(seu.JAK1.002, value = "highlowpJAK1")
# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.002 <- FindMarkers(seu.JAK1.002,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "PROT", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.002 <- filter(markers.002, cluster == "p-JAK1 high")
#markers.002
# Find differentially expressed features between CD14+ and FCGR3A+ Monocytes
markers.002.RNA <- FindAllMarkers(seu.JAK1.002,assay = "RNA", slot = "data", logfc.threshold = 0.3, return.thresh = 0.01, only.pos = T,min.pct = 0.1)
markers.002.RNA <- FindMarkers(seu.JAK1.002,ident.1 = "p-JAK1 high", ident.2 = "p-JAK1 low", assay = "SCT.RNA", slot = "scale.data", logfc.threshold = 0, return.thresh = 1, only.pos = F)
# view results
#markers.002.RNA
markers.002$protein <-rownames(markers.002)
# add a column of NAs
markers.002$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.002$diffexpressed[markers.002$avg_diff > 0.25 & markers.002$p_val_adj < 0.01] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.002$diffexpressed[markers.002$avg_diff < -0.25 & markers.002$p_val_adj < 0.01] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.002$delabel <- NA
markers.002$delabel[markers.002$diffexpressed != "NO"] <- markers.002$protein[markers.002$diffexpressed != "NO"]
plot.vulcano.002min <- ggplot(data=markers.002, aes(x=avg_diff , y=-log10(p_val_adj), col=diffexpressed, label=delabel)) +
geom_point(size=0.5) +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.01), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" adjusted p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 002 min)") &
add.textsize
sign.markers002 <- markers.002$protein[markers.002$avg_diff > 0.25 & markers.002$p_val_adj < 0.01 | markers.002$avg_diff < -0.25 & markers.002$p_val_adj < 0.01]
plot.vln.002min <- VlnPlot(seu.JAK1.002,assay = "PROT",slot = "scale.data", features = sign.markers002, group.by = "highlowpJAK1",ncol = 6, pt.size = 0.5) &
add.textsize
markers.002.RNA$protein <-rownames(markers.002.RNA)
# add a column of NAs
markers.002.RNA$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
markers.002.RNA$diffexpressed[markers.002.RNA$avg_diff > 0.25 & markers.002.RNA$p_val < 0.05] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
markers.002.RNA$diffexpressed[markers.002.RNA$avg_diff < -0.25 & markers.002.RNA$p_val < 0.05] <- "DOWN"
mycolors <- c("blue", "red", "black")
names(mycolors) <- c("DOWN", "UP", "NO")
markers.002.RNA$delabel <- NA
markers.002.RNA$delabel[markers.002.RNA$diffexpressed != "NO"] <- markers.002.RNA$protein[markers.002.RNA$diffexpressed != "NO"]
plot.vulcano.002min.RNA <- ggplot(data=markers.002.RNA, aes(x=avg_diff, y=-log10(p_val), col=diffexpressed, label=delabel)) +
geom_point() +
theme_minimal() +
geom_text_repel(size=2.2) +
scale_color_manual(values=c("blue", "red", "black")) +
geom_vline(xintercept=c(-0.25, 0.25), col="red") +
geom_hline(yintercept=-log10(0.05), col="red") +
labs(x = expression("Log"[2]*" Fold Change"), y = expression("-log"[10]*" p-value"), title = "p-JAK1 high vs p-JAK1 low (t = 002 min)") &
add.textsize
sign.markers002.RNA <- markers.002.RNA$protein[markers.002.RNA$avg_diff > 0.25 & markers.002.RNA$p_val < 0.05]
plot.vln.002min.RNA <- VlnPlot(seu.JAK1.002,assay = "RNA", features = sign.markers002.RNA[1:20], group.by = "highlowpJAK1",ncol = 10) &
add.textsize
plot_002min <- plot_grid(plot.vulcano.002min, plot.vln.002min, labels = panellabels[c(1,2)], label_size = 10, ncol = 2, rel_widths = c(1,2))plot_all <- plot_grid(plot_002min,plot_006min,plot_180min , labels =c("", "", ""), ncol = 1, rel_heights = c(1,1,1))
ggsave(plot_all, filename = "output/paper_figures/Suppl_pJAK1_highlow.pdf", width = 183, height = 140, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot_all, filename = "output/paper_figures/Suppl_pJAK1_highlow.png", width = 183, height = 140, units = "mm", dpi = 300)
plot_all
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_Netherlands.1252 LC_CTYPE=English_Netherlands.1252
[3] LC_MONETARY=English_Netherlands.1252 LC_NUMERIC=C
[5] LC_TIME=English_Netherlands.1252
attached base packages:
[1] parallel stats4 grid stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] png_0.1-7 forcats_0.5.1
[3] clusterProfiler_3.18.1 clusterProfiler.dplyr_0.0.2
[5] enrichplot_1.10.2 org.Hs.eg.db_3.12.0
[7] AnnotationDbi_1.52.0 IRanges_2.24.1
[9] S4Vectors_0.28.1 Biobase_2.50.0
[11] BiocGenerics_0.36.0 ggridges_0.5.3
[13] cowplot_1.1.1 ggtext_0.1.1
[15] ggplotify_0.0.5 ggcorrplot_0.1.3
[17] ggrepel_0.9.1 ggpubr_0.4.0
[19] scico_1.2.0 MOFA2_1.1.17
[21] extrafont_0.17 patchwork_1.1.1
[23] RColorBrewer_1.1-2 viridis_0.5.1
[25] viridisLite_0.3.0 ggsci_2.9
[27] sctransform_0.3.2 ggthemes_4.2.4
[29] matrixStats_0.57.0 kableExtra_1.3.1
[31] gridExtra_2.3 SeuratObject_4.0.0
[33] Seurat_4.0.0 ggplot2_3.3.3
[35] scales_1.1.1 tidyr_1.1.2
[37] dplyr_1.0.3 stringr_1.4.0
[39] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] rappdirs_0.3.2 scattermore_0.7 bit64_4.0.5
[4] knitr_1.31 irlba_2.3.3 DelayedArray_0.16.1
[7] data.table_1.13.6 rpart_4.1-15 generics_0.1.0
[10] RSQLite_2.2.3 shadowtext_0.0.7 RANN_2.6.1
[13] future_1.21.0 bit_4.0.4 spatstat.data_2.1-0
[16] webshot_0.5.2 xml2_1.3.2 httpuv_1.5.5
[19] assertthat_0.2.1 xfun_0.23 hms_1.0.0
[22] evaluate_0.14 promises_1.1.1 readxl_1.3.1
[25] tmvnsim_1.0-2 igraph_1.2.6 DBI_1.1.1
[28] htmlwidgets_1.5.3 purrr_0.3.4 ellipsis_0.3.1
[31] RSpectra_0.16-0 corrplot_0.84 backports_1.2.1
[34] markdown_1.1 deldir_0.2-10 MatrixGenerics_1.2.0
[37] vctrs_0.3.6 ROCR_1.0-11 abind_1.4-5
[40] cachem_1.0.1 withr_2.4.1 ggforce_0.3.2
[43] mnormt_2.0.2 goftest_1.2-2 cluster_2.1.0
[46] DOSE_3.16.0 lazyeval_0.2.2 crayon_1.3.4
[49] basilisk.utils_1.2.1 pkgconfig_2.0.3 labeling_0.4.2
[52] tweenr_1.0.1 nlme_3.1-149 rlang_0.4.10
[55] globals_0.14.0 lifecycle_0.2.0 miniUI_0.1.1.1
[58] downloader_0.4 filelock_1.0.2 extrafontdb_1.0
[61] cellranger_1.1.0 rprojroot_2.0.2 polyclip_1.10-0
[64] lmtest_0.9-38 Matrix_1.2-18 carData_3.0-4
[67] Rhdf5lib_1.12.1 zoo_1.8-8 whisker_0.4
[70] pheatmap_1.0.12 KernSmooth_2.23-17 rhdf5filters_1.2.0
[73] blob_1.2.1 qvalue_2.22.0 parallelly_1.23.0
[76] rstatix_0.6.0 gridGraphics_0.5-1 ggsignif_0.6.0
[79] memoise_2.0.0 magrittr_2.0.1 plyr_1.8.6
[82] ica_1.0-2 compiler_4.0.3 scatterpie_0.1.5
[85] fitdistrplus_1.1-3 listenv_0.8.0 pbapply_1.4-3
[88] MASS_7.3-53 mgcv_1.8-33 tidyselect_1.1.0
[91] stringi_1.5.3 highr_0.8 yaml_2.2.1
[94] GOSemSim_2.16.1 fastmatch_1.1-0 tools_4.0.3
[97] future.apply_1.7.0 rio_0.5.16 rstudioapi_0.13
[100] foreign_0.8-80 git2r_0.28.0 farver_2.0.3
[103] Rtsne_0.15 ggraph_2.0.5 digest_0.6.27
[106] rvcheck_0.1.8 BiocManager_1.30.10 shiny_1.6.0
[109] Rcpp_1.0.6 gridtext_0.1.4 car_3.0-10
[112] broom_0.7.3 later_1.1.0.1 RcppAnnoy_0.0.18
[115] httr_1.4.2 psych_2.0.12 colorspace_2.0-0
[118] rvest_0.3.6 fs_1.5.0 tensor_1.5
[121] reticulate_1.18 splines_4.0.3 uwot_0.1.10
[124] spatstat.utils_2.1-0 graphlayouts_0.7.1 basilisk_1.2.1
[127] plotly_4.9.3 xtable_1.8-4 jsonlite_1.7.2
[130] spatstat_1.64-1 tidygraph_1.2.0 R6_2.5.0
[133] pillar_1.4.7 htmltools_0.5.1.1 mime_0.9
[136] glue_1.4.2 fastmap_1.1.0 BiocParallel_1.24.1
[139] codetools_0.2-16 fgsea_1.16.0 lattice_0.20-41
[142] tibble_3.0.5 curl_4.3 leiden_0.3.7
[145] zip_2.1.1 GO.db_3.12.1 openxlsx_4.2.3
[148] Rttf2pt1_1.3.8 limma_3.46.0 survival_3.2-7
[151] rmarkdown_2.6 munsell_0.5.0 DO.db_2.9
[154] rhdf5_2.34.0 HDF5Array_1.18.0 haven_2.3.1
[157] reshape2_1.4.4 gtable_0.3.0
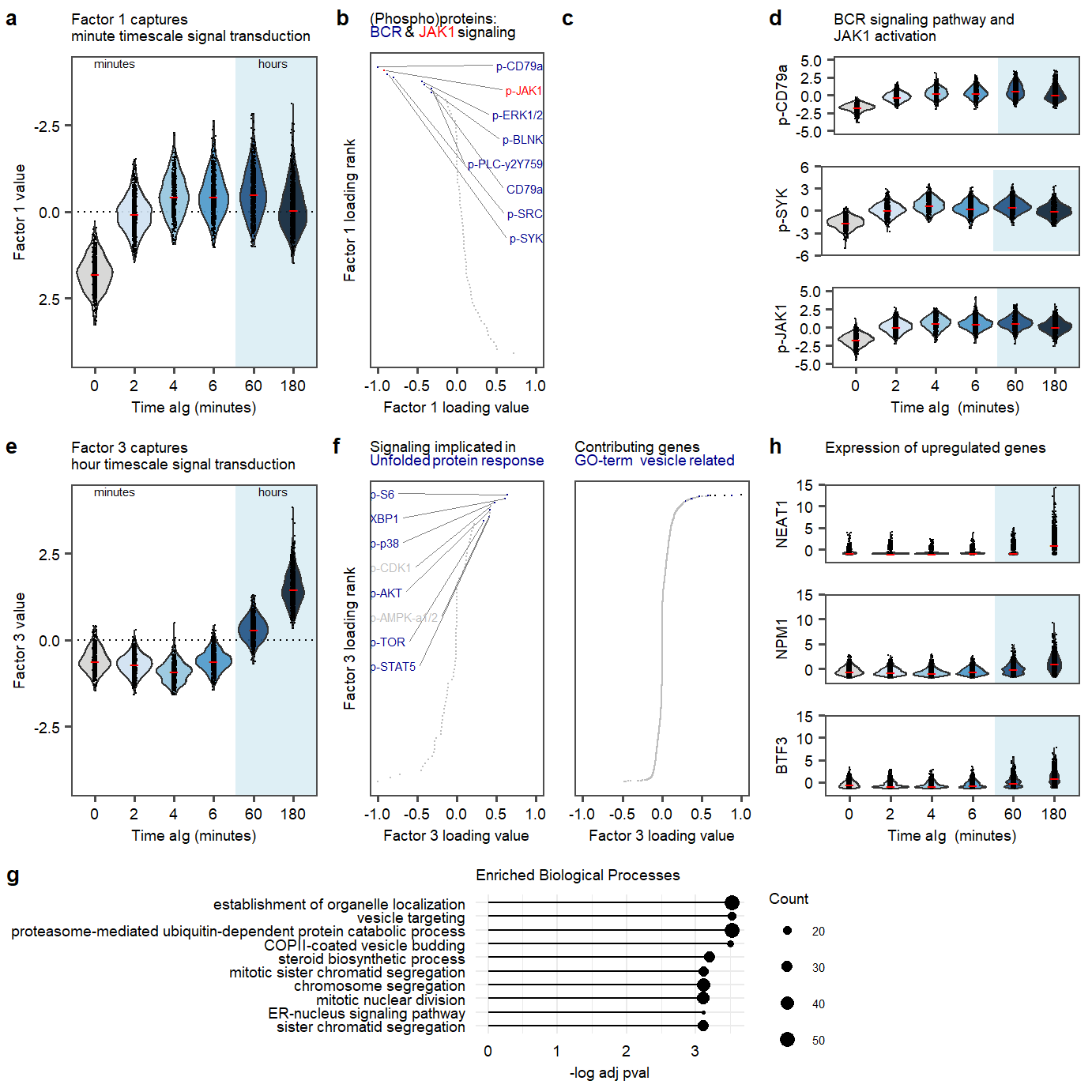 Figure 2. Factor 1 and 3 exploration.
Figure 2. Factor 1 and 3 exploration.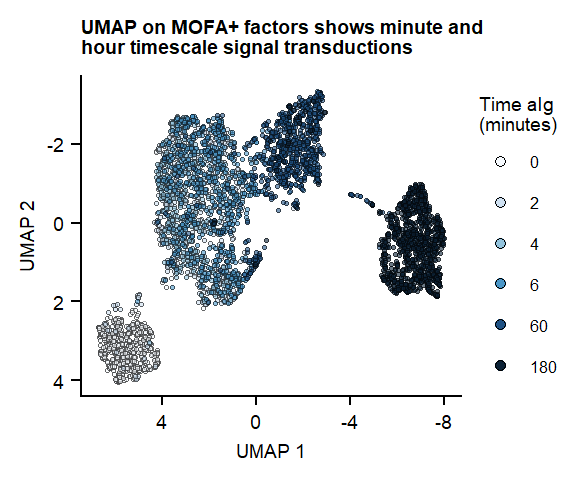 Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements
Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements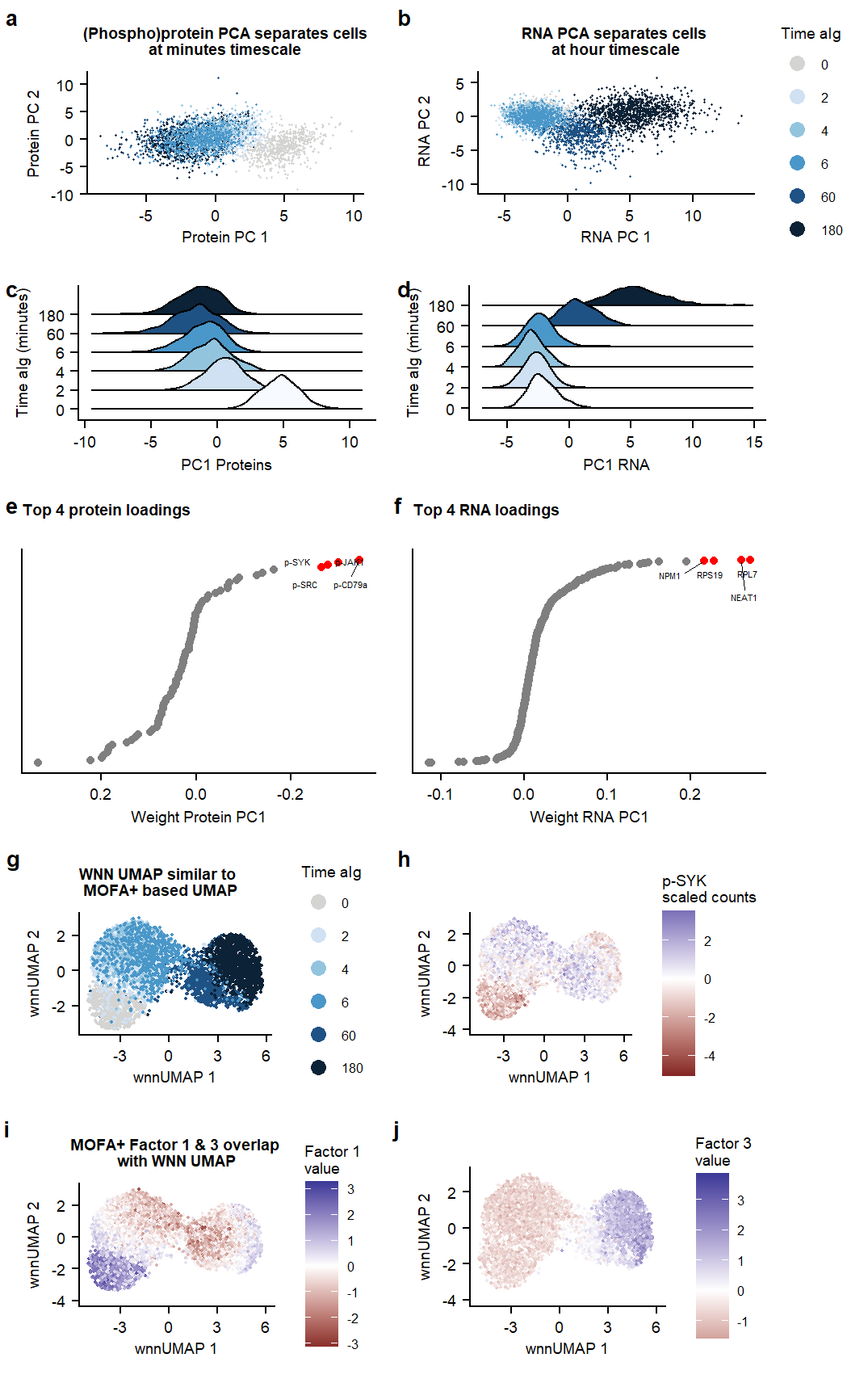 Supplementary Figure. PCA analysis on RNA and Protein datasets separately.
Supplementary Figure. PCA analysis on RNA and Protein datasets separately.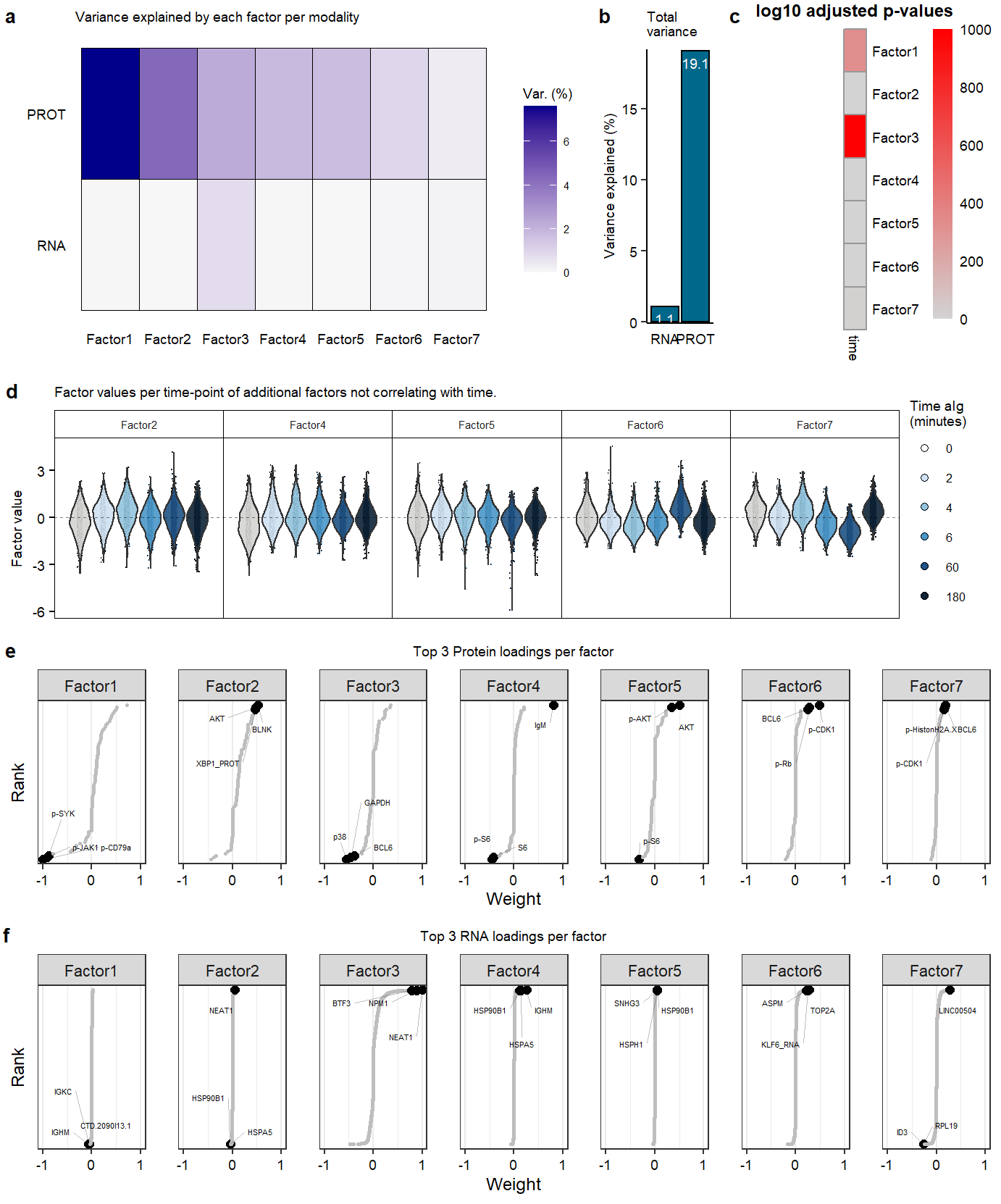 Supplementary Figure. MOFA model additional information
Supplementary Figure. MOFA model additional information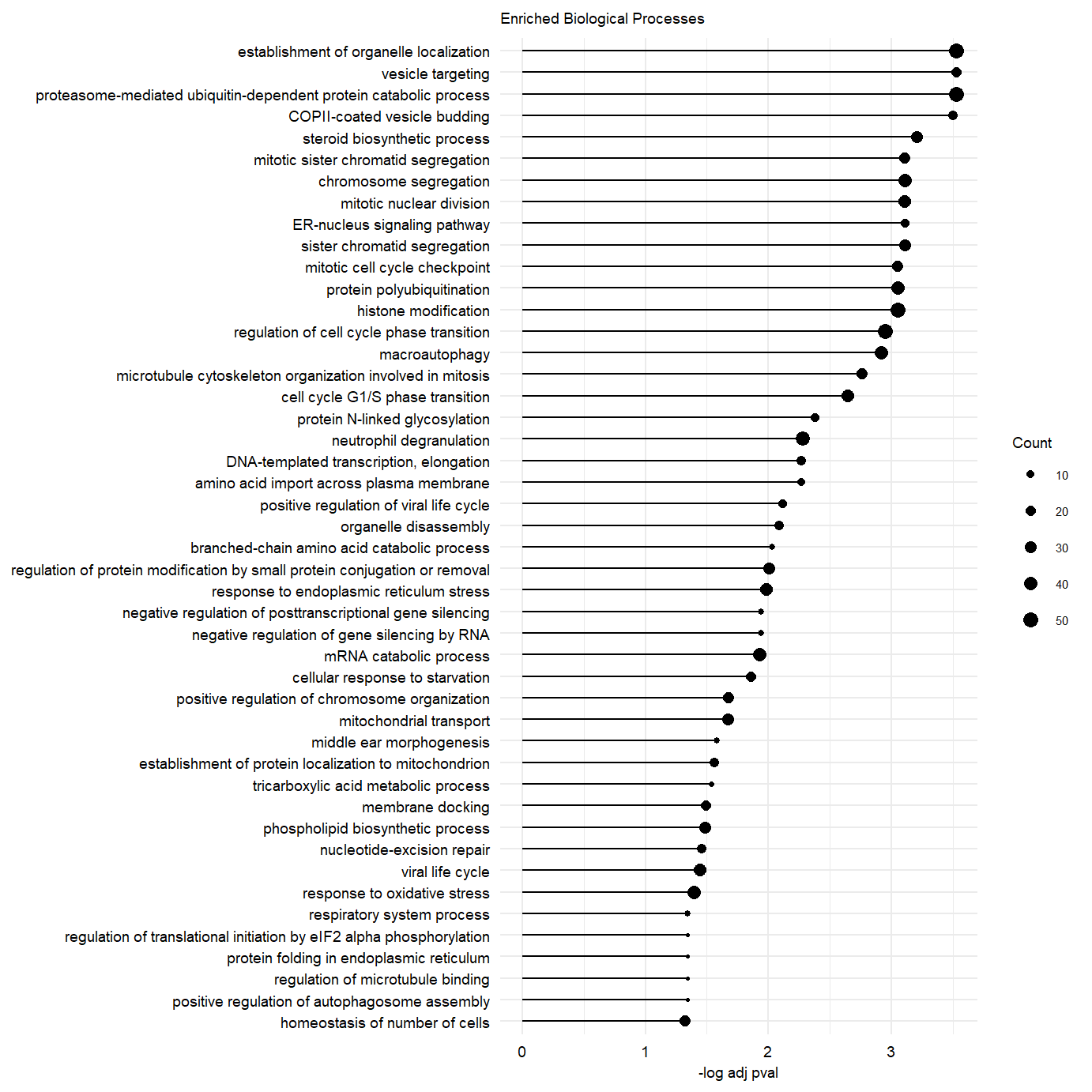 Supplementary Figure. Top 50 enriched gene-sets in postive loadings factor 3.
Supplementary Figure. Top 50 enriched gene-sets in postive loadings factor 3.